I have been meaning to blog the recent CIMI meeting (already 10 days ago 😉 but have been buried in ‘work’. So in lieu of that, I’ll put up an analysis of a real use case from Intermountain Health that Stan introduced on the call last night. Don’t expect to get all the details, but it will give you some feel for what CIMI is doing.
The analysis started with the question “should we allow ENTRY to contain ENTRY in the CIMI reference model”? Obviously my response to that was one of horror, since a ‘clinical statement’ can’t contain a c’linical statement’ …
But that’s just me. Let’s backtrack and get the real reason from Stan as to why this might seem necessary. It’s to do with the need to replicate the typical Intermountain lab panel item CEMs and also construct a ‘panel’ CEM, whose contents would be one or more analyte CEMs. At Intermountain, with their different RM, this is understood colloquially as something like an ‘entry’ (the panel) containing ‘entries’ (the items).
The CIMI (and for that matter 13606 and openEHR) Reference Models are not constructed like this. In these RMs, an ‘Entry’ means a clinical statement (which might contain numerous data points).
Here is my analysis. If this could be perfected, it could help us transform the 6,000 or so Intermountain CEMs into ADL 1.5 archetypes based on the CIMI reference model, thereby making a hugely valuable resource more visible to the outside world.
[Archetypes made for this analysis are here on GitHub]
I used urinalysis analytes and uninalysis panel as the example, since I had those archetypes lying around from last year when I was there.
So let’s consider the logical picture first.
In CEM-land, there are numerous ‘atomic’ analytes that can be considered ‘clinical statements’ on their own. This is because they contain all the context items you would expect, right down at the analyte level. Example – flattened form:
The source form of this CEM is:
You can see that it gets all that context from a StandardLabObsQuantitative CEM, which itself just changes slightly the StandardLabObs CEM.
Now, at Intermountain (all@IHC – correct me if I get this wrong), they build a panel by making another CEM that can include one or more urinalysis analytes. A ‘panel’ is essentially just a collection. Now, the panel CEM will have its own context items, and it ‘contains’ the individual panel item CEMs, each with their own context, like the above. Note that in this approach, the vast majority of context items are repeated in every analyte, even though they logically apply to the panel. (There is a use case that requires overriding some context on a per-analyte basis, but let’s deal with that later).
Now let’s go to ADL + CIMI RM land.
The correct way of modelling the above, in my view is as follows:
- each analyte CEM is converted a CLUSTER archetype containing just the focal items (I thought it would be an ELEMENT actually, but there are two core nodes in each analyte: ‘item’, ‘reference range’. These could actually be collapsed to a single ELEMENT with a DV_QUANTITY in openEHR, because we have an efficient way of representing normal ranges, which saves a huge amount of pain and space, however, that feature is gone from CIMI.
- an ENTRY archetype is constructed to represent the ‘panel’ concept. This can accomodate any number of analytes. It uses a slot to do this.
- now any actually ‘clinical statement’ of the urinalysis variety is always an instance of this ENTRY, with more or less of the required / allowed CLUSTER analyte nodes where you expect them in the panel.
The eco-system I have constructed to do this is partly based on archetypes I built with Joey Coyle and others at Intermountain last year, so it might not be 100% correct according to a re-analysis today, but it allowed me to use all the context archetypes. So the result is as follows:
Where the tree-child relationship = archetype specialisation. So there is
- Panel-level
- a ‘StdLabpanel’ ENTRY,
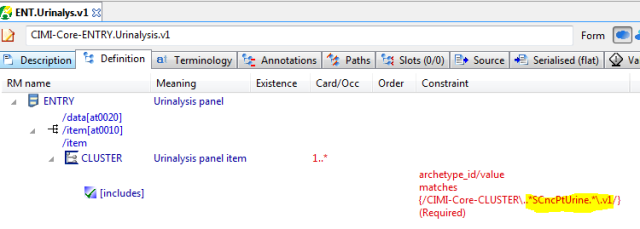
- a ‘urinalysis’ child which limits the possible children to whatever is designated the acceptable set of urinalysis panel analytes (obviously there is more tan one kind of urinalysis, but we are keeping things simple here). Here we tighten the slot to only allow Na and K analytes, because that’s all I have in my example here. Note this slot just says what could appear in the panel items group, not what does.
- a templated urinalysis child, which fills the slot with Na and K analyte CLUSTER archetypes
- I could make numerous different variants of this. You can see how a Chem20 and Chem7 could be made for example.
- Analytes
- you can see at the bottom a lineage of StdLabObs / Quantitative / Na, & K archetypes. Logically these are atomic, but due to the item + normal range thing, they are concretely CLUSTERs.
- Context (quals & mods)
- the middle group under Attribution. These are some of the context items. The remainder are inline in the panel ENTRY archetype. It was done this way (I think) due to reasons of re-use, and it might be that in a better version all qualifiers and modifiers were separated out as CLUSTER archetypes.
So, here’s what it looks like in the ADL Workbench (beta 10, on the point of release ;-):
Urinalysis version, differential form, using the ADL 1.4 style slot, which matches any CEM with ‘SCncPtUrine’ in the name (Intermountain would need to correct this, but you get the idea).
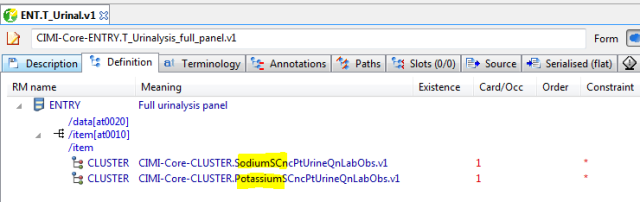
Next, the standard lab obs panel item:
Now the Sodium redefinition of this (flat form):
Now… the whole panel, as a template, differential form:
And finally, the flat form:
This structure is pretty close to what an Intermountain panel CEM would look like for urinalysis.
Note: if I had done another template with just Na in it, then we would have the above, but with just Na instead of Na + K in the core panel items part. So now you can see how we have:
- retained all context (+/- detail errors of mine) for use with either a single analyte-as-a-panel or as a panel with multiple items in it.
- the key is to distinguish a ‘panel’ as the clinical statement here, carrying all the context, from the analytes, which just define the central content, without replicating all the context.
Now, back to Stan’s use case of – what happens if there is a ‘context override’ at a leaf, e.g. Bob Smith did the panel except for Jane, who (re)did the Na analyte (let’s assume this is physically possible with the machine) because Bob stepped out for a coffee. I would deal with this by:
- grouping all the context items in a CLUSTER that you think could possibly have a different value at the leaf level (note: most can’t, incuding everything to do with the sample provenance, etc, also when the specimen was received etc, placer order number)
- including that CLUSTER in the ENTRY for standard lab panel
- also including that CLUSTER 0..1 in a StandardLab item CLUSTER, as a 3rd node (i.e. in addition to the ‘item’ and ‘normal range’).
That’s more or less what is already happening in the current CEMs (i..e the inheritance I referred to at the top).