happy new year and best wishes for 2014. I hope your new year’s day is a bright one (unless you live in the UK, in which case it’s a lost cause here today 😉
I have been working in the last few months to produce a final version of ADL/AOM 1.5, based on:
- existing requirements,
- emerging requirements – Intermountain, CIMI,
- Harold Solbrig’s proposals for terms-as-URIs,
- Dave Carlson’s MDHT / AML work at OMG led by Robert lario,
- general feedback on the openehr-technical list, particularly from David Moner’s group at UPV, where they have implemented different rules
- implementer feedback
So here’s the new idea. To date, We have been trying to keep ADL/AOM 1.5 backwardly compatible at the syntax level for ADL 1.4. However, I think this keeps too many old problems unsolved. I propose a new approach:
- make the central ADL/AOM 1.5 specifications as clean as possible
- provide a series of updates to ADL 1.4, coming from the 1.5 specs, that are carefully designed to be applied to 1.4 tools, to bring them up to date
- e.g. things like how to post-fit the new identifiers, tuple support, annotations, to DAL 1.4 archetype tools
- provide rules and tooling to deal with differences between archetype paths, upon which querying is based
- provide a 1.4 => 1.5 upgrade tool to completely convert existing ADL 1.4 archetypes to the new format
The latest changes I propose (and have in fact implemented) are primarily about dealing properly with the long-running problem(s) of archetype node ids, i.e.:
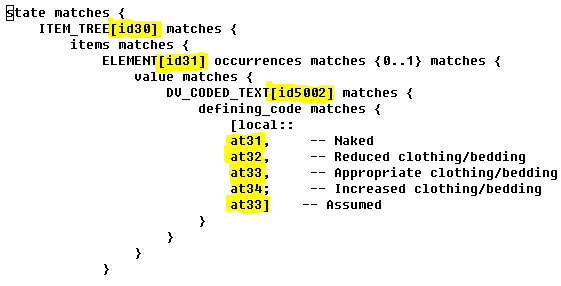
- the quirky 4-character top-level codes like at0001, at0003 etc, also lower level codes like at0001.0.1, at0.0.2
- the root node code at0000, which really should be ‘1’ not ‘0’
- the question of whether all nodes should have codes or not (in openEHR we have not done this; the 13606ers have)
- the lack of separation between node id codes and value codes
- sibling alternate C_OBJECTs under a single-valued C_ATTRIBUTE were not properly distinguished by their paths
The proposal is documented here on the wiki.
All comments and criticism welcome. If you think the proposal is broken in some way, or could be done better, don’t be afraid to say so. Please comment on the openehr-technical list, or for substantive comments, the wiki page above.
Let’s try and get to a final proposal that works for all ADL/AOM users – not just openEHR. I think that would be a real achievement.