One of the age-old debates in health informatics: can there be ‘one information model’ for shared clinical information? Some dream of a model to rule them all, uniting standards efforts, while others dismiss the idea as impossible or unrealistic. Obviously inside deployed health/hospital information (and all other – lab, GP, nursing, billing, PAS etc) products, there are private, differing information models. These do not concern us.
What is of interest is the information model upon which shared information is based. ‘Shared’ being any information exported from a source system to a receiver, or to a shared EHR. This information is the usual target of standardisation. To date, numerous health information standards have divided up this world into segments – messages, EHR Extracts, ‘documents’ (a meaningless world, but nevertheless…), and so on. Each of the half dozen or so standards bodies have produced their own model for one or more of these segments. The combinatorial space contains some dozens of information models. As soon as we consider the fact that core clinical, administrative and demographic information is essentially the same, whether it is sent as a ‘message’, a ‘document’ or an ‘extract’, it is clear that having differing and competing information models is non-sensical. After all, why should there be multiple definitions of a ‘microbiology result’? Clinicians only see one… multiple information models just means needless data transformation.
Objectors to a ‘single information model’ say ‘whose model’? Or ‘models embody different points of view’, or that that noone will ever agree on one model because humans are just plain bloody-minded. Well, they sometimes are…. but I think the real reason for this stance is that noone has produced a convincing argument for a standardised model. Let me try that now.
In order to have any hope of agreeing a ‘single information model’, we need to agree a general architecture, since this defines the role of the information model. Then we need to be precise about the function of the information model in this architecture. For the argument here, I will just use the abstract openEHR architecture, shown below.
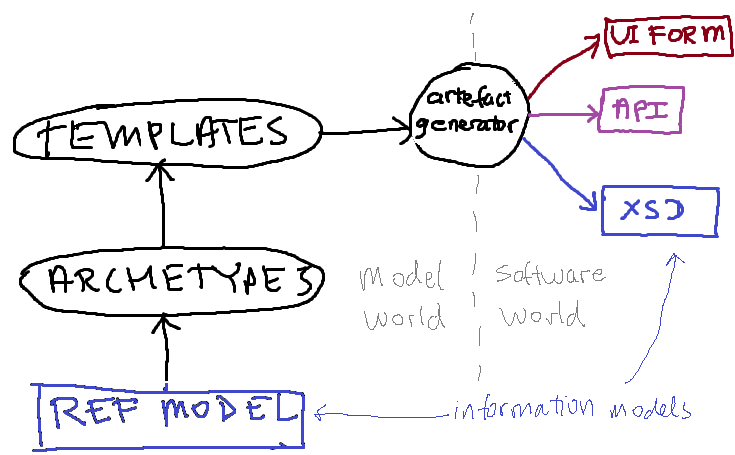 In this architecture, an information model designated the ‘reference model’ (RM) is the basis for defining ‘archetypes’ and ‘templates’. Adherents of HL7v3 messages or CDA will understand ‘archetypes/templates’ to be something like *MIMs and ‘HL7 templates’ respectively (the exact relationship between the RM and archetypes/templates in openEHR is not identical to that in HL7, but the general sense is the same).
In this architecture, an information model designated the ‘reference model’ (RM) is the basis for defining ‘archetypes’ and ‘templates’. Adherents of HL7v3 messages or CDA will understand ‘archetypes/templates’ to be something like *MIMs and ‘HL7 templates’ respectively (the exact relationship between the RM and archetypes/templates in openEHR is not identical to that in HL7, but the general sense is the same).
Now, in openEHR, templates (a kind of archetype) are used to generate downstream artefacts, such as UI forms, APIs (i.e. source code), and data schemas, typically in XML schema (XSD) format. The latter is also an information model, but a specific one, e.g. a microbiology result model. There is a mathematical relationship between any such downstream ‘information model’ and the RM, via the archetypes and templates.
The first obvious question now is: which information might we try to agree on – the RM, or one of the specific XSDs? Agreeing on the latter would be practically useful, and might be possible socio-politically, since each such model is of a limited piece of content, such as a particular lab result, investigation, observation, diagnosis etc. But people are only likely to agree to this if they are convinced that such models are generated by an architecture they can also agree to.
So we need to ask the question: what is the function of the reference model in this architecture?
The first purpose is to act as a semantic basis for developing the next layer of models – the archetypes. Since archetypes are constraint models, they can’t invent structural semantics of their own. But they need structural semantics, as required by the numerous actual types of clinical content. For example, an Apgar result requires some notion of time (there are always at least 2 samples), and some notion of a list of ordinal values, with a sum. A glucose tolerance test result requires a notion of time, some idea of patient state (fasting, post glucose challenge), and a group of values. These requirements can be thought of as patterns. Here are a number of key patterns that are known from building hundreds of archetypes and other models in health informatics:
| PATTERN | EXPLANATION |
| + data / state / protocol (/reasoning) | In observation data, separate out data (actual datum being recorded e.g. BP) from patient state (e.g. lying, standing) and protocol (cuff type, instrument type) |
| + History of events | Provide a structure containing 1..* Events, allowing data and patient state at each one, supporting intervals, point events, and math functions, e.g. ave/delta/max/min |
| + Tree structure | Generalised free-form tree for containing clusters of data items, e.g. the 5+1 Apgar items, numerous microbiology result items, etc. |
| ++ Order state machine | A way of recording current state in progression through a standard state machine applying to any ‘order’ |
| ++ Composition / document | An aggregation concept acting as a ‘bucket’ for information recorded by a professional at a given time for a given subject of care. |
| + Participation | A pattern defining the connection between parties (people, organisations, devices) and other information. |
| +++ Party / role / accountability | A pattern defining relationships between parties, including those that are roles played by some underlying actor. |
There are more, including patterns to do with security, versioning, identification, etc.
A reference model containing these patterns enables them to be used directly in the next layer of content models (the archetypes, templates, etc); without them, each archetype has to re-invent the structure in some simpler pattern (typically a simple node-arc tree structure as used extensively in HL7 RIM-based models). Now the idea of ‘agreeing a single information model’ is really the idea of agreeing on these patterns. It seems reasonable that only one ‘history of events’ pattern could be defined and used throughout shared health information. After all, accountants agree on double-entry book-keeping (a kind of data structure) and logicians and electrical engineers on truth tables (another kind of data structure). Indeed, if these patterns were explicated and made an object of standardisation, I believe that some would be agreed quite quickly. Some however, are trickier, particularly the party/role/accountability one (entire chapters in books by Martin Fowler and Len Silverston, to name two, are dedicated to this). The ‘+’s in the table above indicate my idea of how hard each pattern would be to agree.
Now, even if we assume that difficult patterns such as party/role/accountability create furious debate, it doesn’t seem impossible that some agreement could be found. After all, the last 10 years of e-health standards activity has involved much furious debating, and to be frank, only very modest outcomes – including the numerous aforementioned incompatible models. No real attempt has ever been made to try to agree on ‘patterns’, but this is the essence of what needs to be standardised in a ‘reference model’ in the above architecture.
Incidentally, the main way we developed the patterns used in openEHR was not to get a bunch of IT specialists into a room to argue about UML diagrams. Instead, we built a draft of the RM and then clinical experts tried to archetype it. Where they ran into problems was an indication of something wrong in the RM, which we duly adjusted. After some years, the RM became stable, and in its current version 1.0.2, is extremely solid, and safe to use for industrial software.
Which brings us to the second purpose of the RM – implementation in software. Clearly in this role, the exact form is likely to be an augmented version of the RM needed for the purpose of archetyping. As long as this augmentation is done carefully (e.g. with wrappers, no changes to existing attributes etc), then the instances created by the software will be consistent with those constrained by archetypes and templates, and also, with data created by the various downsteam artefact types.
Agreeing on this augmented RM is likely to be harder, and may not be useful; indeed, there might be more than one augmented RM, e.g. for messaging, for EHR and so on. But agreeing the limited version does not seem such a tall order: agree on what core patterns are needed, and then agree on the formal expression of each of those patterns – using the content model proving approach used in openEHR.
Why has this never been done? Because (to my knowledge at least) no-one ever posed the standardisation problem in this way before. It may not be not too late…

Pingback: ICMCC News Page » One information model to rule them all?
Pingback: Interoperability Requirements #3: Information Structures | Health Intersections Pty Ltd
Aaah. My RIM. My precious.
Cross linking to my own comments on The One Model Falacy
One model to rule them all will only come about when we agree
a) on viewpoint, and b) on objective, measurable criteria for inclusion in the model.