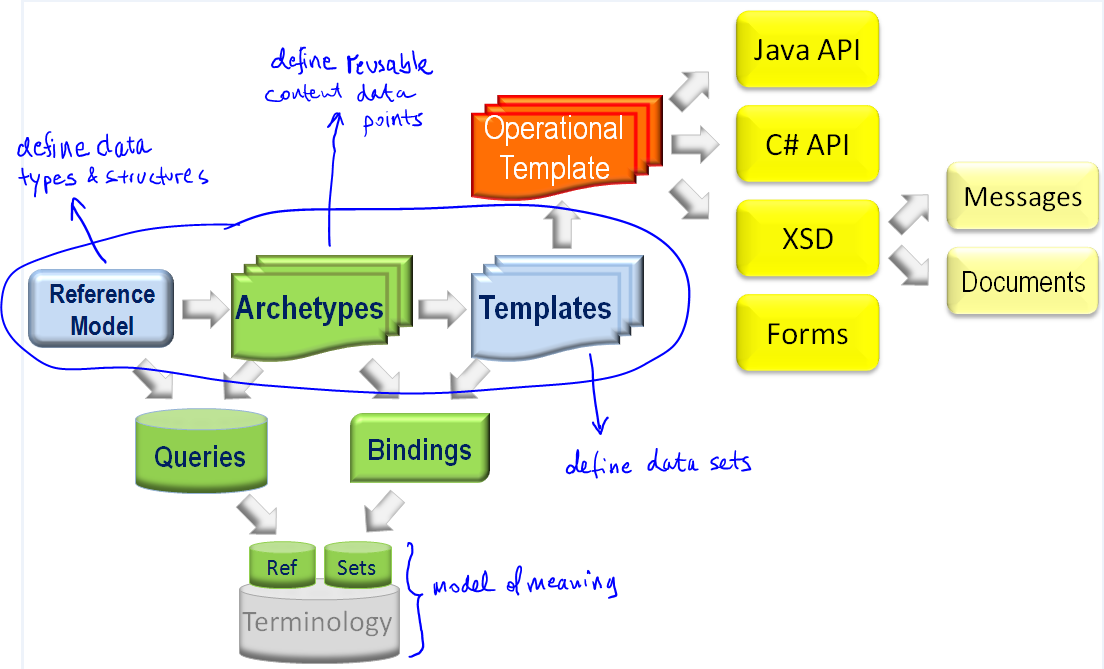
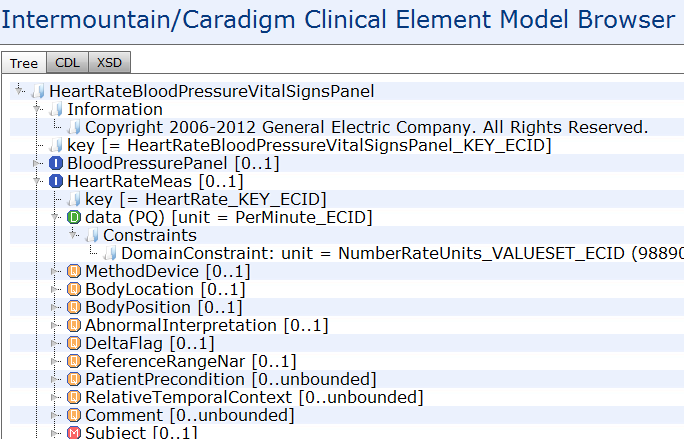
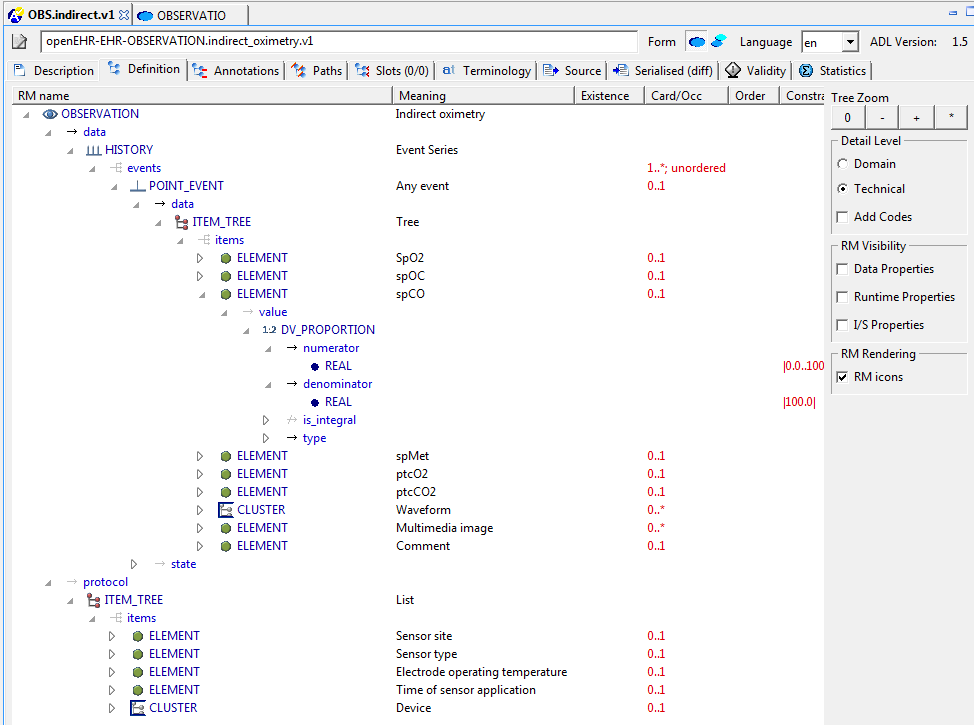
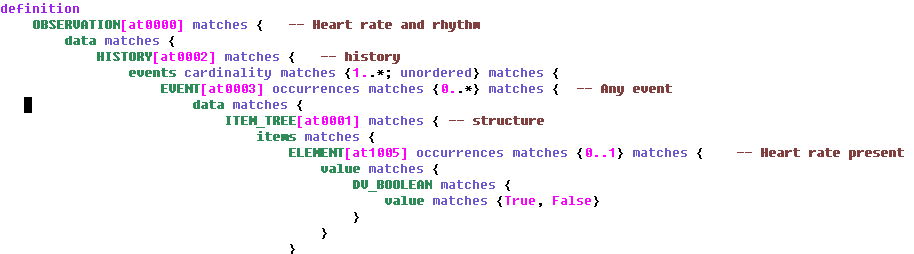
Following the DCM meeting convened by Dr Stan Huff (Intermountain Healthcare) in Washington in July, reported in an earlier blog post, there is a further meeting this week in San Diego, which will discuss the issues of ‘data types’ and ‘reference models’ for the purpose of DCM (detailed clinical models).
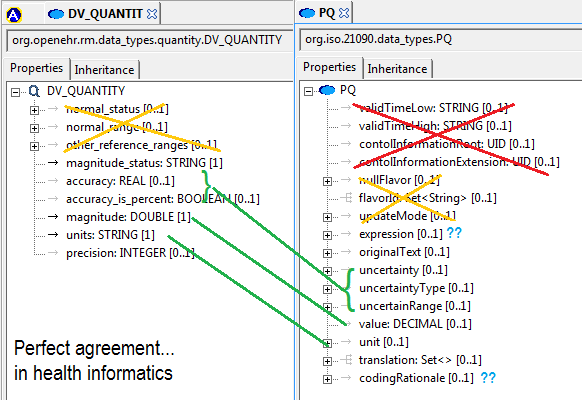
I created two slideshows to explain my views on these matters (DCM_and_data_types and DCM_and_reference_model [both PDF]). Below is an extract of my arguments in these slideshows, based on experience, for adopting a particular approach to data types and reference model within the stated mission the DCM forum, which is to find formalism and attendant models in which to express universally shareable detailed clinical models. Naturally, my view on ‘the answer’ to that question is ‘openEHR (ADL/AOM) archetypes, templates and terminology’, but what I am providing below is not an argument supporting that, but one proposing how to proceed with respect to the ‘underlying models’.
Continue reading →