
The above shows a typical web form calculator for the CHA2DS2-VASc score, used for estimating the risk of stroke in patients with non-rheumatic atrial fibrillation (AF), primarily for the purpose of deciding the use of anti-coagulant therapy [Wikipedia].
How would such a score be programmed?
The thesis of our approach in openEHR is the use of a high-level guideline programming language, which you may think of as an updated version of things like Arden Syntax. It aims to be more comprehensive and to solve some of the problems that Arden does not.
Here’s how the core calculation looks in the current version of openEHR Decision Logic (see the full module here):

The above calculates the basic score. There are further tables commonly used to convert the raw score to a stroke risk, such as the following.
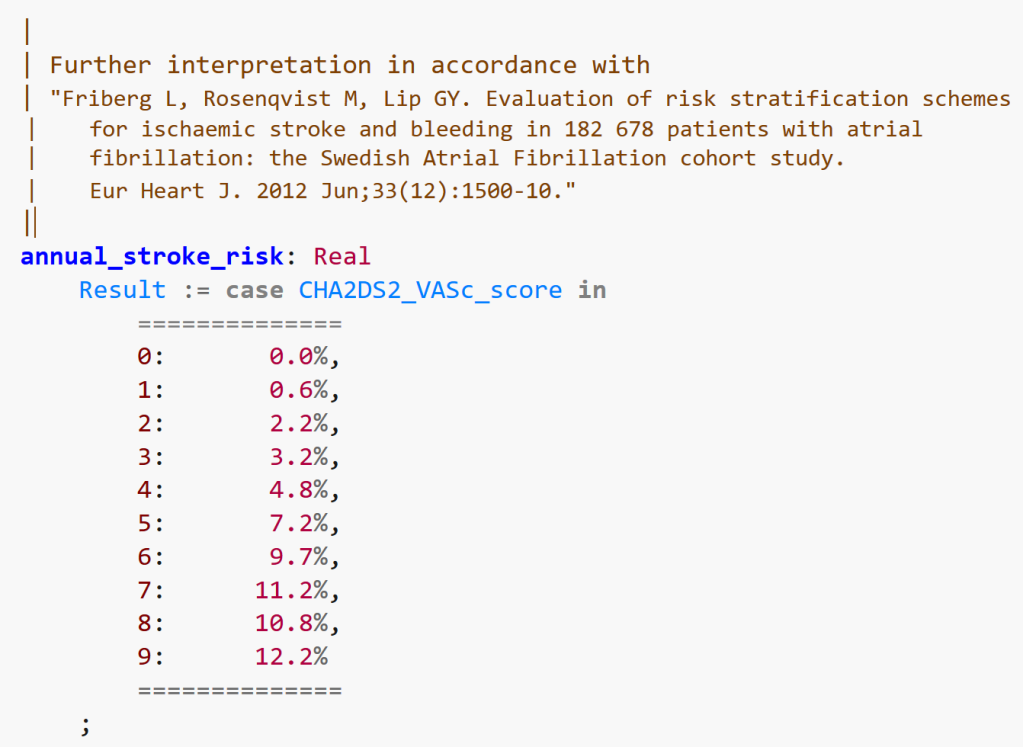
The question here is: can we envisage that with a bit of training, domain experts (in this case, cardiologists, experts in AF, anti-coagulation etc) and clinical informaticists could use this kind of high-level language? Assume that there is a nice text editor with auto-complete, syntax colouring and so on.
The above syntax – openEHR DL – has been designed so that it appears as close as possible to the published (and indeed, mental) form used in the domain – in other words, to provide a cognitively easy pathway to converting (or authoring de novo) natural language guidelines in computable form. openEHR DL is integrated with related formalisms representing Plans as well as subject proxy variables, rather than being standalone, so it may be an attractive direction for the future.
With a published meta-modal and grammar, decision logic modules (DLMs) written in a language like the above could become an attractive way to share computable clinical guidelines.
Making such a language attractive means including features, even syntax tricks (such as the long line comments to create the appearance of in-line tables), to make this kind of programming appear natural to clinical / medical experts.
Over the next few months I will publish more such snippets of well-known guidelines in openEHR DL format, including some other interesting tricks.


You surface two common motifs seen in EHRS (and other applications): domain specific language for clinical rules and assessment instrument/questionnaire/survey/scale (you plug data in and some result pops out. Risk assessment, severity scores, and prognostic instruments are common.
The biggest problem w/ a clinical domain specific language has been the data query. This goes beyond Arden syntax’s well known “curly braces” problem.
* The author/developer needs to know the names of the data elements they want to use.
* How do you deal with conflicting entries in a chart? I recall one patient I needed to figure out if they needed antibiotic prophylaxis. I heard a heart murmur. I dug through the chart looking to see if someone had addressed it before, echo reports, cardiology consult. I found recent exams which reported a different murmur from what I was hearing. But, I don’t know how certain the author was about their finding.
This raises another issue: how to express certainty which separates the uncertainty that something exists (I.e. I know I hear a heart murmur) vs. certainty about correctly identifying the specifics (I.e. the patient is too tachycardia to tell if systolic or diastolic).
I also found notes which said “no murmur”.
Which do you select as being the truth (in reality, everything in a medical record has some probability of being in error, as well as clinician judgement being subject to cognitive errors, judgement and knowledge limitations.)
* How do you know that the data element you found actually fits the criteria needed for a rule/tool? Many, if not most, such instruments have some notion of what fits and what doesn’t. E.g. has_heartfailure: does DOE, rales and pedal edema count? Abnormal EF w/o clinical manifestations?
This is of particular importance if you also add factors like severity. SNOMED, for example, let’s you qualify findings/disorders w/ severity (mild, mild-to-moderate, moderate, moderate-to-severe, severe, life-threatening (child of “severe”), and fatal.
There are standard definitions for grading severity for a lot of disorders, but it’s a pain to find the guideline or review paper which publishes such.
I am an emergency physician, so I may have a different perspective on severity (e.g. of an allergy) compared to someone who works primary care. For me, anaphylactic shock, upper airway angioedema requiring treatment, and significant bronchospasm spasm (e.g. not relieved by 20mg of albuterol given over an hour). I.e. crap that requires a procedure to maintain airway, as well critical care. A provider not used to seeing this spectrum of disease may think wide spread hives as severe.
There is the US NIH (maintained by NCI) CTCAE which have specific criteria for grading severity. All of the disorders/findings in CTCAE are in SNOMED (I did a mapping a few years ago, and got the “life threatening” concept addd to SNOMED). There is a fairly standard definition of what makes a disease mild, moderate, severe, so the severity of disorders is roughly comparable between disorders.
I like CTCAE and think we need it in EHRS, NOT just clinical trials.
* The rule/instrument you are using may have very specific definitions of the data which may not be discernible in your EHRS. A common error I have seen is the indiscriminate use of LOINC codes which are part of a survey/questionnaire/assessment scale/clinical decision rule. An end user may assign a data element a code that has what seemed like a good fit, but in reality has a lot of “baggage” (specific definition, exclusions which are part of the instrument, etc).
* You need to know the range of values a data element might take in order to understand it. You may need to convert from an ordinal scale to nominal. You might need to compare multiple values and write an algorithm to pick a conclusion (e.g. hopefully has_heartfailure is a conclusion which is specific to the assessment instrument/rule/etc.)
Obviously we want to enter data just once. Adding a check box for “has_heartfailure”—or any US CMS “quality” indicator—is a fail on the part of the EHRS developers if the answer is already in the EHRS. If they cannot find it/use it, they should hire a qualified informaticist who can show them how it should be done.
We need to understand how important manually entered data really is. We need better ways to get all the details entered into a single note authored by patient, their family/care giver, nurse, and physician. Keeping track of who entered what is essential, but at the end of an encounter there needs to be a single source of truth. There are a lot of issues and details, but it’s time to migrate beyond our paper-charts mentality.
Most EHRS (last four enterprise systems I have used in particular) totally devalue my skill at collecting a H&P, expressing nuance and uncertainty, but still make me the most expensive data entry clerk, making me spend 1/3 to 1/2 of my shift fighting w/ an EHRS rather than taking care of my patients. They are inefficient, lack built-in decision support, and you end up with narrative text which is expensive and error prone to extract non-trivial information from. Phooey.
Designing an EHRS in 2021 that is based on paper charts and forms is about as smart as designing a self-driving electronic vehicle based on a stage coach. Obvious to say so, but I am sure you have seen this anti-pattern crop up over and over. Health information is complex, and a lot of that complexity was handled poorly by paper charts and how we were trained to “write a note.” At least w/ paper charts you could draw diagrams, sketch anatomy, etc. My iPad works fine for that, just not a lot of EHRS out there which can treat data as computable information and let you draw.
Back to my main point, we need better data in EHRS, esp. the CC, HPI, ROS, PE, medical decision making, diagnostic considerations, procedure narrative, and plan of care. SOAP notes are a thing of the prior century, when adding chart dividers and a problem list were super helpful. Since the invention of the SOAP note we have seen the rise of EHRS (invention of cell phones, rise of the internet, mandatory use of seatbelts, etc).
If we get enough data about some given data element (hopefully informed by assessment instruments/etc) with the explicit understanding that we don’t know all the ways information will be used at the time we enter it.
So, in summary: (1). Getting data into the chart with enough detail and qualification (including how precise, certain, and ambiguous a given finding/history of/diagnosis) is a current problem which need a change in how we think about what is an EHR. (2). Data in an EHRS may, or may not, be suitable for use in standardized assessment instruments/scales/scores/rule. (3). Most EHRS are a total dogs breakfast on the back end so figuring this out turns into most of the effort in writing a clinical rule.
Solution: rules based expert systems need to drive the user experience, and we need to abandoned the out-dates notion of what a health record is. An EHRS, in the 21st century should reflect human factors engineering and cognitive science best practices, not dead tree based technology.
Hi Kevin,
good to hear from you. Yes, we are very aware of the whole data access problem, and indeed you are pre-empting later posts. The first thing is that each DLM has a way to declare its variables locally, in a clean form, e.g.
systolic_blood_pressure: Real
currency = 2h
;
This would also be documented right there with the specific description of what is meant by ‘systolic blood pressure’ in this instance. If it is what is usually meant, i.e. systemic arterial BP measured measured at systole, within the last 2h, then it should say so.
Similarly variables like has_congestive_heart_failure have to be defined to say exactly what they mean. If the meaning of ‘has CHF’ here in one DLM is not the same as in some other, you might name the variable as has_chf_cha2ds2vasc or similar.
Now, how do all these variables get connected to their data? The short answer is that they are managed in a dedicated Subject Proxy Service, which contains bindings for all such variables to the back-end systems available in the current environment. This approach has the effect of separating Decision Logic Modules from the bindings to a particular HIT environment. It is in this service that you would write the appropriate query or API call (it may be a whole routine) to obtain exactly the flavour of CHF or anything else you need. See – https://specifications.openehr.org/releases/SM/latest/openehr_platform.html#_subject_proxy_service_sps