
Some readers may have read my previous post FHIR compared to openEHR. If not, I recommend you do, it is available in Spanish, Japanese and Chinese as well as English. Here I aim to clarify some of the concrete differences which are increasingly common sources of confusion, particularly with the FHIR hype wave preventing coherent thinking in many places. It seems that the human psychological pre-disposition for uncritical silver bullet thinking is as strong as ever, but I still hope (perhaps vainly) that in e-health we can soon get back to real science and engineering.
Design Intent
The design intent of FHIR is to enable applications to extract data from opaque systems for a) applications (so-called B2C), and b) other systems, i.e. messaging (aka B2B). Why ‘opaque’ systems? Firstly, nearly all commercial HIS and EMR systems today are proprietary and their vendors contractually control data access. Secondly, non-opaque systems – those containing data based on published standards and with open access to legitimate users – can export their data in their native form, via standard technology. Such systems don’t need another model imposed through the APIs or messages – indeed doing so creates risks for errors and patient safety.
FHIR’s modelling approach is to create definitions called profiles, based on resources, which are fragment definitions. Obtaining a blood pressure in FHIR means designing a profile based on the use of the Observation resource, and then implementing a REST API that extracts that kind of data from each system you want to get it from (Cerner, Philips, InterSystems, Allscripts, VistA etc). The FHIR ecosystem looks as follows.
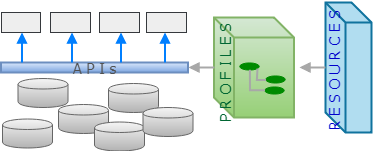
openEHR’s design paradigm is an architecture for a health information platform, complete with REST APIs, information model, domain modelling layer (archetypes and templates), portable querying and generated resources for interoperability. Its information model (aka Reference Model) is designed to represent health information content, context, audit, and versioning, allowing applications to commit data to a coherent longitudinal EHR, as well as read and query it. More can be found here in my earlier post openEHR technical basics for HL7 and FHIR users. The openEHR ecosystem looks as follows. (Note, for both cases, I have not included terminology in the diagrams, for the sake of simplicity).

What is the difference between retrieval fragment models and an information capture model? There are two main differences:
- factoring: fragments are typically factored with all possible ‘context’ data (audit, timing, other meta-data) on the fragment, since this is a convenient way to retrieve a focal element (like blood glucose) without having to know all the containment structure in which it was committed (different in every system), and do half a dozen separate queries to get all that context. Consider the FHIR Observation resource. The data elements basedOn, status, subject, focus, and encounter will in general apply to a number of observations and/or other information items persisted in a given encounter or other health system contact. A commit model designed on this basis will contain replicated attributes all through the hierarchy, instantly creating the standard data representation problems that go with replication.
- semantics: fragment models (like FHIR resources) don’t constitute a coherent ‘model’ of semantics as such, rather they are models of isolated lumps of information, obtainable in an ad hoc way. This is a perfectly reasonable design approach for ad hoc data retrieval. FHIR resources are designed based on inspecting data available from legacy systems and creating a superset that covers 80% of them (the 80/20 rule). A good example is the ProcedureRequest resource, which is pretty clearly a ‘bag of attributes’. The openEHR RM and TP models on the other hand are fully designed models of clinical, demographic and planning information, with underpinning theory.
Modelling Methodology
Could FHIR modelling have been done another way? From the point of view of openEHR, the answer is yes. Currently health data in FHIR are modelled as ‘superset’ resources, with profiles being used to reduce / constrain and add elements, to make up most of the domain specifics. This means FHIR doesn’t have a core library of models that are clinically ‘designed’ (even if some downstream profiles might be). In openEHR we do the opposite; clinical and informatics professionals design archetypes according to principles and needs, not what data are inside their (unloved) EMR systems. And yet the most common use of archetypes today is to define data coming from existing systems – precisely the use case of messaging and FHIR. How can that work, you say? Not complicated – openEHR uses a ‘recombination’ layer of models called Templates to define data sets, which are used as message definitions for legacy systems, as well as data sets for new applications, including forms. See this post on openEHR technical basics for HL7 and FHIR users to see how this works.

This methodology is sometimes called the ‘maximal data set’ approach, because the archetypes are not directly models of anything, but definitions of possible data points and data groups. This approach has been in use in industry for over 10 years, and the archetype formalism was made an ISO standard in 2008, and again this year (or next, depending on when the final completion occurs). If HL7 used the archetype / template distinction, we would have been able to work on a global standard library for clinical data (or better – if they had used the archetype formalism). This was also the long-stated intent of CIMI, now an HL7 working group. Going by recent communications I see, CIMI will be limited to being a slightly more abstract version of FHIR, or perhaps nothing at all. A great pity.
The result of all this is that FHIR is creating a de novo ‘non-designed’ set of clinical data models in competition with much larger and more established designed model bases, such as openEHR CKM (7500 data points), the VA FHIM (at least some thousands) and Intermountain Healthcare’s own Clinical Element Models (a model base with 20 years’ of history, and 7500 clinical data points). This will not help the global e-health effort. A major missed opportunity.
Data Representation
In openEHR, all EHR data are instances of a stable Reference Model. This enables stable software and DB schemas to be constructed from the 150 classes of the RM. Similarly, data representing Task Plans (another 80 or so classes) are instances of that model. Since all clinical and other domain level models are in a different layer from the information models, addition of new domain models doesn’t affect the information model level. This means all openEHR data around the world, from Moscow to Leeds to Seoul, are automatically interoperable at the information level, and where archetypes are shared (the evidence so far is that 70% of the 500+ CKM international archetypes are routinely re-used), they are interoperable at the domain level.

It’s not immediately clear what would be considered the ‘data model’ of FHIR, but the most obvious candidate is a sub-set of the resources. The FHIR resources are at very different ontological levels, from infrastructure (StructureDefinition), primitives (data types), to the general (Observation), to the clinically specific (Specimen, BodySite, AdverseEvent). I have not been able to discern any rule for when a new resource is created versus when a profile of an existing resource is created. Should ‘Skin Review’ be a profile on the Observation resource, or a new one? Why is MedicationRequest a resource, and not a profile of a Request (or Order) resource? Because it contains a continuum of ontological levels, the FHIR resources have two characteristics undesirable in a stable information model:
- volatility – clinically specific resources will clearly need to change over time – how this affects dependent profiles is an interesting question;
- open-endedness – one has to presume that the set of resources will simply keep growing to accommodate new major clinical information categories.
The consequences of these factors are that the model is ‘never finished’ and that any databases or software based on it will likewise need continued maintenance. For a model of data extraction, this probably doesn’t matter too much, but anyone trying to work in the other direction will need to think carefully about the consequences.
Domain Coverage
This is probably the area of most concern to anyone who is hoping for interoperability over the 50,000 data points of clinical information we are likely to need over time (excluding genomics and proteomics data). The openEHR CKM and other CKMs provide models for about 10,000 domain data points today (an average of about 14 per archetype, approx 700 archetypes).
Currently FHIR has some hundreds of data points, mostly in the form of resources. About 1/3 of these correspond in some way to the classes of the openEHR RM. The rest – possibly 300-400 – address the same domain space as the 10,000 openEHR data points. This represents a 20-30x difference in coverage. There are however some profiles published (e.g. simplifier.net), which although immature, one presumes are intended to try to cover the domain space (interestingly, this site claims to have 5439 profiles and 27,043 ‘resources’, the latter of which is hopefully an error – if not, FHIR is in trouble).

If organisations using FHIR want to do that, they need to either replicate the work of openEHR and/or just use resources in a generic fashion. Actually creating thousands of clinical model data points means creating a large clinical modelling community, something like openEHR has done, and/or committing serious long-term resources as Intermountain Healthcare and the VA have done. It also requires solid modelling tools like those used in openEHR, as well as a high-quality formalism, such as the archetype technology created by openEHR.
The question of domain coverage will be covered in the next post in detail.
Building Profiles v Archetypes
In openEHR, all clinical content and other domain definitions, such as Task Plans, are defined as archetypes and templates based on the Reference Model (main model of EHR content), or another stable base model such as Task Planning. Archetypes and Templates are expressed in the Archetype formalism, consisting of the ADL language and Archetype Object Model (AOM). Archetypes can be specialised, composed and templates enable further specialisation and terminology binding of included archetypes, locally to the template.
In FHIR, definitions of clinical data might already be resources (e.g. Medication, CarePlan), or they may require profiling (e.g. Observation). The resource library is a mixture of domain- and technical-level models, with the former existing in various levels of generality. Consequently there are resources for things like ClinicalImpression and ProcedureRequest (archetypes in openEHR of the Evaluation and Instruction classes, respectively), as well as Observation (in openEHR this is an RM class). One of the downsides of building resources for clinical specifics is that those models will inevitably need to change over time. If you engineer say Java classes based on those resources, you are up for software changes; if you have any database (say, caching FHIR responses), the schemas will also need to be changed over time. With any serious amount of deployment, these will cause problems.
To create a specialised version of a resource, you create a profile, and/or extensions of a parent resource. To create say ‘blood pressure’ in FHIR, you will create a profile based on the Observation resource.
The following provides some idea of how blood pressure looks in FHIR and openEHR. The first image is from simplifier.net BP profile, and shows ‘hybrid’ view of the profile and resource. As can be seen, most of the Observation resource attributes are inherited, with nodes for systolic and diastolic pressure being added at the end, in a generic tree of BackboneElement nodes. This exposes one of the odd things about the Observation resource: its design is oriented to representing a single principle data point such as a patient weight or blood glucose, hence the valueQuantity and dataAbsentReason nodes at the top level. But the general case is that clinical data occur in groups (e.g. blood pressure, physical exams, and many others). Consequently, to represent blood pressure, the top-level valueQuantity node is removed, and the necessary value nodes are added in the Component sub-tree.

The following is the openEHR CKM blood pressure archetype, first shown in the ADL workbench, which is a technical tool providing the view equivalent to the above. As can be seen, it contains significantly more domain elements (although presumably FHIR could match this just by adding a bigger ad hoc tree at the bottom of the Observation resource), as well as a time-series data structure. The lack of the latter in FHIR means that 1000 vital sign samples in one openEHR Observation would require 1000 separate FHIR Observations, each repeating the numerous context attributes. The openEHR data items are also separated into the categories data, (patient) state and protocol, enabling more precise processing.

The following view shows part of the archetype in a technical mode, showing underlying RM classes and attributes (the equivalent of the greyed names in the right hand column of the FHIR model).
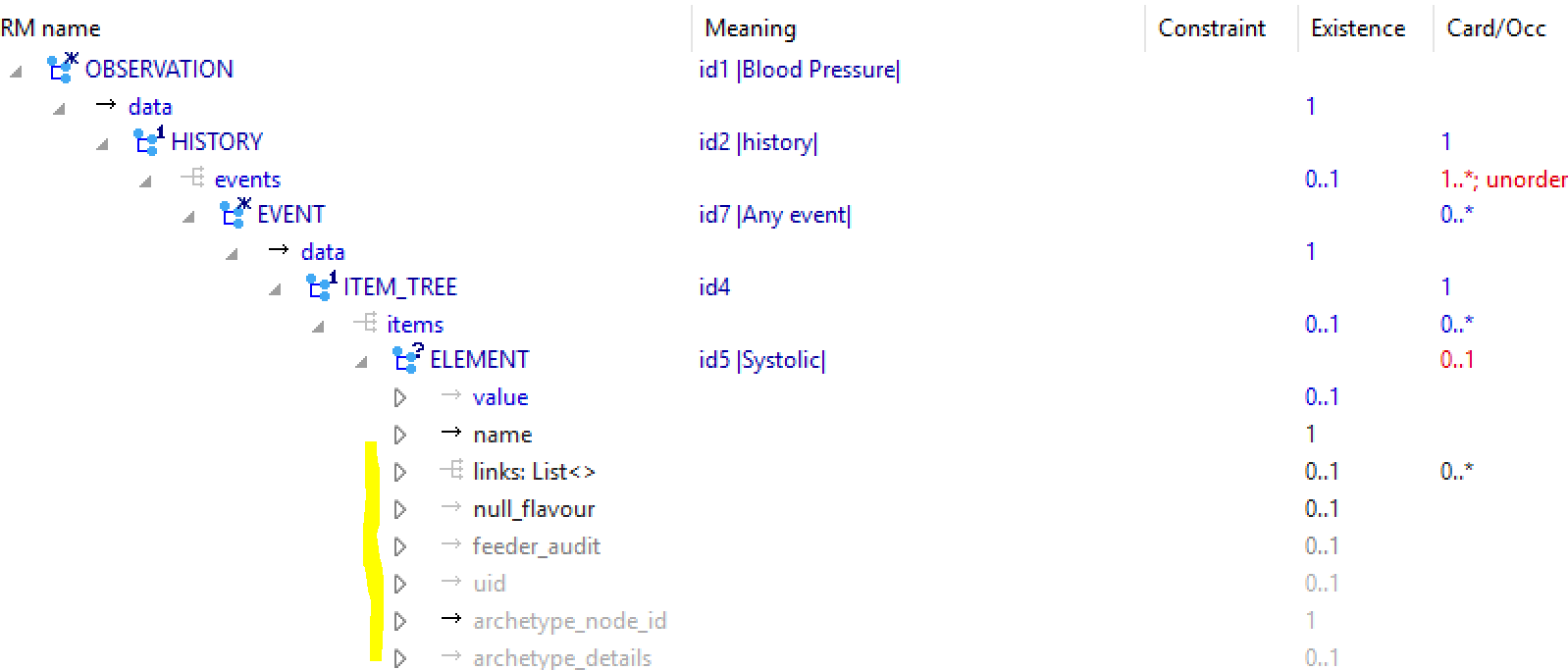
The following view is from CKM, and is one of the views clinical modellers normally use.

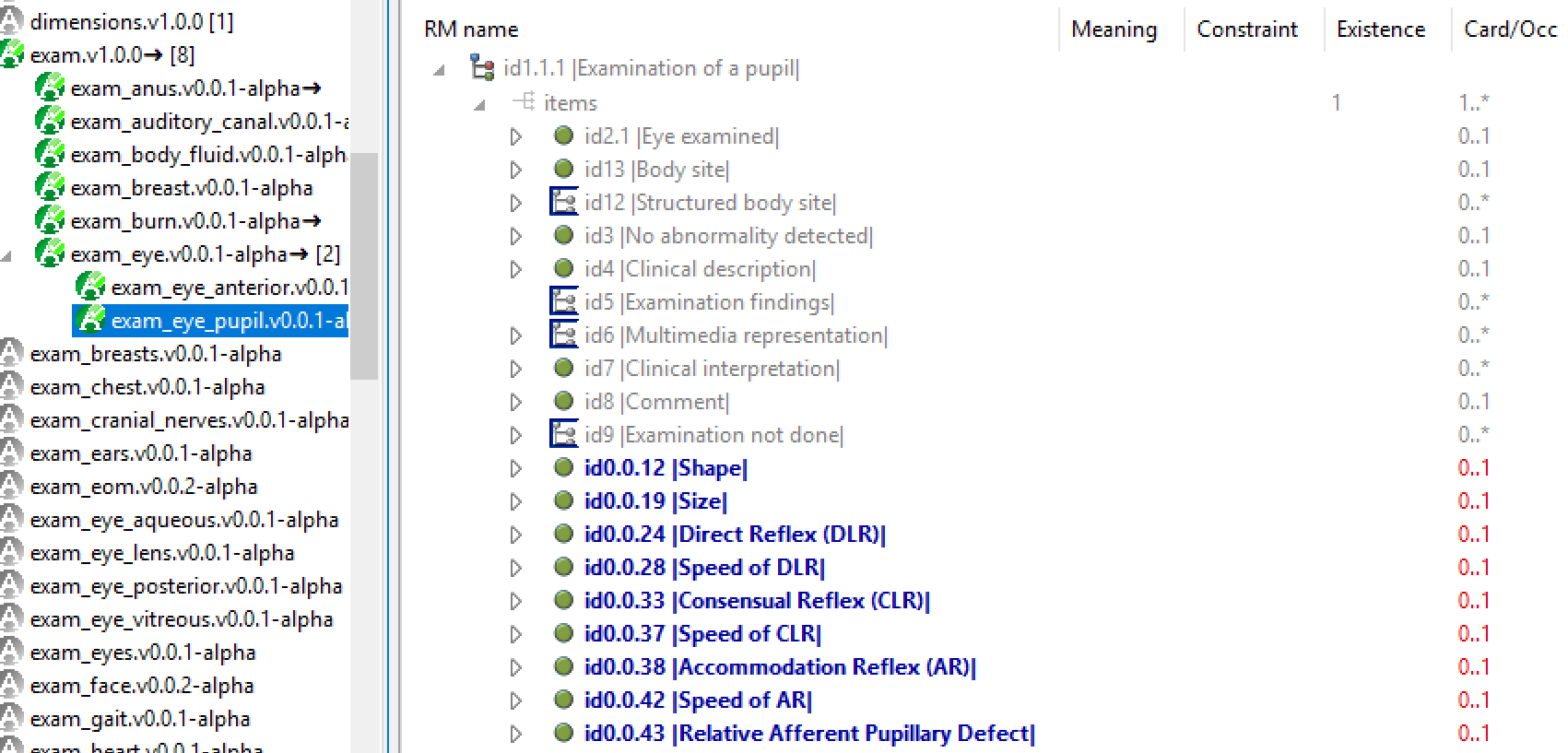
Archetypes can be infinitely specialised. The following shows the CLUSTER archetype eye_exam_pupil, a specialisation of eye_exam, itself specialised from exam. In the inheritance-flattened view, added and changed components are visible. The key property of specialisation in openEHR is that queries based on the top level archetype will work with data created by any specialisation. This is because tools implement strict conformance checking all the way through the inheritance hierarchy.
It’s not clear yet whether this kind of formal modelling will work in FHIR, which is based on the StructureDefinition resource.
It’s also not clear how FHIR works in Chinese, Russian, Spanish, or Farsi.

Some common misconceptions
Could FHIR be used to implement an EHR?
I have to admit to being amused at this question, which resurfaces from time to time since HL7v3 and CDA, earlier HL7 standards for message and document sharing respectively. Perhaps the people who ask such questions just don’t read what is on the tin; one has to assume they have also never thought about what it takes to create an EHR. Certainly the attempts to do so with HL7v3 and the CDA were not successful.
To take the question seriously: what prevents a message model from being used as a model of committed data in an EHR? As noted above, fragment definitions are factored for retrieval, not commit, and secondly (at least in the case of FHIR), because it is not a semantic model of a committable structure but a superset model of existing system contents. An information capture model is a fully structured model of context and meta-data based on specific design principles reflecting the semantics of the data – EHR, demographics, etc. But building an EHR doesn’t stop there. One needs bullet-proof versioning (a major feature in openEHR), digital signing, an order state machine, and advanced features such as process plan representation.
A shared EHR also needs semantic querying, which is to say querying based on the domain content models. For example, a way to query systolic BP values created by different applications in different data sets over time (perhaps to evaluate a stroke risk guideline). Implementing semantic querying requires a language like Archetype Query Language (AQL) and a modelling framework that enables every item of data to be semantically and automatically tagged in deployed systems (done in openEHR via archetype node ids). The following shows an AQL query. Being based only on archetype identifiers and paths means the query is portable across systems using the archetypes, regardless of physical storage or DB.
SELECT obs/data[id2]/events[id7]/data[id4]/items[id5]/value/magnitude, obs/data[id2]/events[id7]/data[id4]/items[id6]/value/magnitude FROM EHR [ehr_id/value=$ehrUid] CONTAINS COMPOSITION [openEHR-EHR-COMPOSITION.encounter.v1] CONTAINS OBSERVATION obs [openEHR-EHR-OBSERVATION.blood_pressure.v1] WHERE obs/data[id2]/events[id7]/data[id4]/items[id5]/value/magnitude >= 140 OR obs/data[id2]/events[id7]/data[id4]/items[id6]/value/magnitude >= 90
Can openEHR be used to implement Interoperability?
Of course, we’ve been doing that for over a decade. Just like any other well defined model-based framework in any domain, the data structures needed for interoperability are machine-generated from the base models. In fact this has been done in other domains since the days of rpcgen, a Unix tool from 30 years ago (think Google Protobuf, but not object-oriented). Why standards organisations are still manually building wire definitions for healthcare is a mystery. In openEHR, the only message definitions are generated from models. Currently, openEHR uses XSDs called Template Data Schemas (TDS), generated from Templates by tools including the Template Designer and ADL-Designer. Other messaging technologies may be targetted in the future.
openEHR also REST APIs, as the basis for applications to talk to the EHR and other services. In the future we may see interfaces for Apache Thrift, Avro, and Google protobuf, and GraphQL among others.
The real priorities: Pathways and Process
For the majority of clinical people who are saddled with the routine inability to share data even between two instances of the same vendor product in different hospitals, constantly failing referrals and missed hand-offs, more Interoperability, i.e. point-to-point communications between proprietary components and applications, is not that exciting (even if it is how our lab data and prescriptions are generally managed). Most other data remain unshareable, and current approaches don’t even touch the main problems for providers:
- process: the inability to define and track process of care of a patient through the healthcare system prevents any meaningful realisation of continuity of care.
- computable guidelines: much of healthcare is driven by guidelines and care pathways, but these are still defined and consumed as documents, with no IT support at all, despite growing complexity.
The hierarchy of concerns is as follows.

To date, Interoperability has been tinkering with the bottom (patient data) level and mostly failing, other than for a few well defined kinds of message. And the reality is that no amount of Interoperability will be able to extract a picture of patient process (the second level) from existing systems, because there is simply no coherent representation of process available to be extracted. By definition, a picture of patient process has to be defined in an open way and work across existing systems, not be hidden inside them. As for the top level, non-computable guidelines are the best we have are either PDFs, e.g. Intermountain Healthcare (to take just one good US example with which I happen to have some familiarity) or web pages e.g. UK NICE. These represent very high quality evidence-based medical research, but are entirely processed by humans.
So, although FHIR (and to be fair, HL7v2) can enable applications to do some nice things with data from other places, it won’t get us much closer to a solution for patient-centric, process-based healthcare, for the simple reason that the problem cannot be solved inside proprietary systems or applications that talk to them.
The open e-Health Platform
To achieve true continuity of care that includes computable pathways, guidelines, process management, and shareable data, will require a patient health record outside of proprietary systems. This will need to be conceived of as a platform, of the kind described in the UK Apperta Foundation white paper. There is no reason why this cannot be used alongside existing and new products, the so-called ‘bi-modal approach’. The primary challenge is achieving the appropriate separation of patient-centric from application-centric models and data.
For anyone who wants to achieve true continuity of care, driven by care pathways, the bimodal environment is the primary target. The different approaches of FHIR and openEHR address different needs in this environment, and their usage will change over time as the balance shifts to a shared process-based continuity-of-care platform.
Addendum
Some readers will take some of this post as being critical of FHIR. I have been critical of a few elements in a scientific sense, in the spirit that I would expect any ideas to which claims for fitness for use are attached to be critiqued. Indeed, FHIR has some nice things, like the URI system and the terminology approach. However, we should all be critical of unsubstantiated claims, that is to say hype, which is the fake news of the tech sector. Hype is noise from the mouths of the irresponsible intended for the ears of the gullible. The hope of promulgators is that the audience will turn off its critical faculties and believe that a single blood sugar displayed on a screen can be linearly extrapolated to the e-health platform of the future. My 24 years’ experience in e-health tells me that the vast majority are too intelligent to be fooled in this manner. Hopefully we can prevent those who control budgets and e-health plans from falling into the traps of previous waves of silver bullet thinking.


Excellent piece Thomas! As you so rightly point out, the real priorities are pathways and process -where we are now and where we need to be. I hope we get there, and soon. Look forward to reading the UK Apperta Foundation white paper.
Great!
Permanent link to Chinese (zh-CN) translation of this valueable post:
FHIR v openEHR – concreta (Table of Contents)
https://mp.weixin.qq.com/s/ScBtqtEEzXz3m3zBg430nw
Thanks for your insights 🙂
Holiday season greetings to you and your loved 🙂
Best Regards,
Lin Zhang