Ler en Español (traducción – Diego Boscá Tomás)
日本語で読む(Shinji Kobayashi による翻訳)
中文 (Lin Zhang)
I see a growing number of organisations and individuals posing the old standards comparison question, today, in the form of: how does HL7 FHIR compare to or relate to openEHR?
It’s always fun to revisit this question from time to time, especially as some of the questioners are fortunately young enough not to know too much of the history of e-health standards. To understand the question requires looking at a few basic health informatics concepts. Below I’ve tried to do this as objectively as possible, but of course I know more about openEHR than FHIR at the detail level.
How is the need for interoperability solved?
The traditional answer, and the basis of HL7, EDIFACT etc is to say that we can’t control what is in systems, so we need to impose standards on data that flow between systems, i.e. messages. That worked successfully for many years for certain well-defined data, particularly lab results and prescriptions, and indeed, many systems are still pumping out millions of HL7v2.3 or v2.5 messages as I write this.
However, I would argue (and I believe most of the openEHR, 13606 and HL7 CIMI communities would agree) that:
- in the last 20 years, the range of semantics and data that need to be shared to enable distributed e-health is far wider than can be accommodated by messages that capture specific sets of data, and in fact the size and complexity of these semantics is such that it is simply unsustainable to have them described either inside specific vendor products (as screen forms) or even specific message formats, meaning they must be re-expressed for each new format (UI display, mobile data capture, various document formats, etc) and technology (various kinds of DB, Javascript etc);
- unlike 20 years ago when many solutions were just screens over database tables, builders of systems today are very cognisent of the semantics they need inside;
- the people who understand the semantics of the domain are not IT people, they are domain professionals and health informatics experts with deep domain knowledge (a recent post illustrating this).
My view is that the only scalable way to create the semantic specifications is for them to be artefacts outside of both vendor products and outside of specific communications formats. This implies a single-source model-based approach, with the models existing in their own formalism, tools and community. Examples of ‘models’ include terminologies, e.g. ICDx, SNOMED, ICF etc; openEHR archetypes and templates, and those of other groups like HL7 CIMI, computable guidelines, drug dictionaries and so on. These various kinds of models (including those of openEHR) have two key characteristics:
- they are created directly by people who understand the semantics of the domain;
- they can be machine converted into any reasonable format, including message formats.

This post on semantic scalability provides some figures on the need for single-source, grass-roots approach.
The message theory versus the system theory
The above approach might be called the ‘system’ theory of interoperability: semantics are formalised by the domain and then machine-converted to concrete forms (e.g. XSD, JSON-based screen formats etc) usable for by systems and tools, and also for messages or other serialisations.
In the message theory of interoperability, message content (not just generic structures or comms protocol) is standardised and becomes a/the conformance requirement for systems, or at least for some of their communications channels. This is potentially fine from the point of view of at least some consumers of data from those systems, but for systems that want to export their native or model-based semantics, it presents somewhat of an obstruction. This is because to communicate on a pre-standardised message channel, they have to engineer conversions between their data and the structures required by the messages. These may be quite different and the mapping may be non-trivial – and wherever there is mapping there are risks to performance, correctness and patient safety. If the message data sets are small and simple, there will be little problem, but if we widen the need to be able to ‘communicate anything’, then it may be a serious problem.
However, we still need to be able to serialise data and send it places. The ‘systems’ approach to doing this is to define the models first – for us, openEHR templates, which include terminology linkages – and to machine-generate message definitions from these. This is a trivial operation today, and in fact has been since the days of RPCgen. Consequently, model-based eco-systems don’t need messages, only generic messaging technology. Note that under this approach, there is not only one single format possible: it may be any kind of generic XML, JSON, binary, or even document formats like PDF, openDoc etc.
So under these two theories of interoperability, the models are in two different places. In the ‘systems’ approach, which openEHR uses, they are outside products and concrete communication formats; in the message approach they are within a particular communication format.
Doing it the systems way is technically harder, but more scalable and more future-proof. Doing it the message way produces quicker returns, but will run into issues when the format needs to change or the size of the content space grows too large.
More on the limitations of the message approach in this recent post.
The ideal world – universal models
An ideal approach for the world of e-health would be to develop a universal set of clinical models (as before, ‘models’ includes terminology, subsets, archetypes, templates, guidelines etc) and tools that can turn them into artefacts usable for building systems components, databases, messages and so on. This ideal was clearly articulated by Dr Stan Huff at the inception of CIMI in 2011. It’s been our thinking since the ate 90s and Stan’s, at Intermountain Healthcare, forever as well. (CIMI has since become an HL7 technical committee, but I believe still has the same aims).
In an alternate reality, the approach to ‘doing messages’ would have been to ‘do models’ and then generate any kind of message or other serialisation format to suit specific circumstances.
However. International standards politics, differing needs, timelines and many other pressures have prevented this, even though it is technically possible and implemented, just not universally used.
I would add that the ideal way to do this kind of work is not via design-by-committee as found in old-style SDOs, but via engineering, performed in a broad open-source style of project (or a number of cooperating such projects). This is the approach we take in openEHR.
The real world
In the world we actually live in, things are more complicated. Firstly people are trying to solve different problems. One problem – statistically undoubtedly still the most common – is to get data out of opaque systems that don’t want to give it up (generally due to contractual / IP rights reasons) and that have limited proprietary interfaces or even just raw RDBMS tables. Another one is to build a new generation of systems and components that embody a modern understanding of healthcare semantics, i.e. information, processes, business rules, CDS etc. These challenges correspond to distinct approaches.
In the e-Health standards world, HL7 ditched the HL7 v3 effort after 15 years (too complex, not well adapted to modern technology, HL7 v2 was working pretty well) and started the FHIR effort in about 2012.
We in openEHR continued with the ‘systems’ approach described above. The related EN 13606 and more recent CIMI communities have worked on a similar basis, as have, tacitly, IHTSDO, drug DB publishers, various guidelines issuers, etc (i.e., these organisations produce artefacts whose content is independent of vendor products and communications formats).
How does FHIR compare to openEHR?
The primary intention of FHIR is to solve system-to-system (B2B) and system-to-application (B2C) communications, without making assumptions about the systems. The B2B part is essentially the message approach for the 21st century; the B2C part involves building APIs suited to programming and addresses a major need – making it easy to build modern e-health applications. The challenge is that the latter clearly starts to make assumptions about what underlying systems’ business logic and content includes.
The primary intention of openEHR is to solve the patient data challenge – long-lived, versioned, distributed and computable patient records – and to do so in a future-proof, semantically enabled way, using a platform architecture. We see interoperability as a natural technical outcome of a framework based on formal semantics. Of course, openEHR itself just proposes some elements of such a platform – it needs to be used with terminologies, ontologies, drug databases, service interfaces and so on.
Technical Representation
FHIR is defined in terms of a library of ‘resources’ (web-data micro-formats) and a technical approach to ‘profiling’ and ‘extension’ to support localisation (these terms are all defined on the FHIR site). In other words, there is some level of ‘clinical modelling’ going on, but it is inside the FHIR XML master formalism. The resources are designed to be instantiated and accessed over REST APIs.
In openEHR, we use a standard Reference Model, and various layers of models on top, including archetypes, templates, and terminology subsets. The openEHR Reference Model is standard for the whole world, and all openEHR data, no matter where they are, obey it. In openEHR and other ADL-based communities, the models are outside the messaging or web micro-formats and are instead machine-converted to any of those forms as required. This clearly means more tooling: e.g. tools to generate basic XML, JSON, Google protocol buffer format, whatever it may be. But the reality is that these concrete preferences keep changing – in the 5 years that FHIR has been running, the main preference of many already evolved from XML to JSON.
openEHR archetypes are represented in Archetype Definition Language (ADL) / Archetype Object Model (AOM, an ISO standard). ADL is also the representation of the HL7 CIMI models. It supports composition, specialisation and redefinition, enabling adaption of more general models into more specific and localised forms.
There are also differences in the style of models being built. In openEHR and related technologies, they are generally built by communities of clinical domain experts, based on requirements i.e. ‘what we would really like’. In FHIR, the stated strategy is to produce each FHIR resource as a model of a superset of data found in legacy systems, and to directly cater for developers’ needs. This leads to different results – FHIR models are directly usable by developers, but are unlikely to be easily re-usable in different technologies, e.g. generating UI forms. openEHR uses tools to generate the XSD, JSON, Xforms, and other formats that developers may want.
Further, in openEHR we have a fairly extensive theoretical basis for the EHR, which informs the Reference Model on which they are built. I am not aware of any equivalent in FHIR. I think this will matter significantly when it comes to interoperating clinical process across e-Health infrastructures.
There is more on openEHR for FHIR afficionados in a recent post, openEHR technical basics for HL7 and FHIR users.
Platform coverage
If we agree on the general idea that a technical computing platform consists of information models, semantic models, APIs and querying, we can make some further comparisons.
openEHR has extensive coverage of patient information (its Reference Model defines numerous kinds of structures), significant coverage of the semantic models area (we think perhaps 25% of general medicine), and limited coverage of the application API area (example), although with some nice tooling, not yet standardised. This is one of the main areas we are working on in 2017, along with new process specifications (currently, a new Task Planning specification is out for comment).
FHIR’s coverage on APIs is substantially greater, as this is what it majors on, and indeed the FHIR project could probably lay claim to having done ground-breaking work on how to engineer APIs for complex data and interactions.
What its coverage of domain semantics and information models is harder to determine – it depends on what one thinks FHIR resources are – something like archetypes, or a kind of Reference Model, or something in between? My reading is that they are like a library of openEHR templates, with re-use and specialisation available for extention, plus a set of generic resources for identifiers, data types, generic structures, demographic resources and other infrastructure types. One difference between the two is that a more complex resource, say Risk Assessment, is its own data structure, whereas in openEHR Health risk assessment is an archetyped form of the RM Evaluation class. Adding any new clinical content in openEHR is just a case of adding new archetypes (i.e. no change to the Reference Model); in FHIR it requires new resources, or else profiling of some existing resource. This implies new software for each new resource.
One area I believe openEHR has covered well is semantic querying, for which it uses a portable query language, AQL. To my knowledge, FHIR doesn’t have an equivalent of this. Portable queries are powerful because they are independent of physical storage schemas and technology – they only depend on the archetypes.
The different coverage of essential platform elements tends to lead to one or other eco-system being more immediately interesting to different types of developers, but a lot of what to use and how comes down to what problem one is trying to solve.
Hype versus Reality
It is probably worth briefly mentioning the problem of ‘hype’ if only to alert the unwary. FHIR has been heavily hyped, and many would say over-hyped, as per this industry piece. This isn’t to point at the FHIR project, but rather to a combination of the standard human psychology of the silver bullet (the irrational belief that one solution will solve all problems) and a certain amount of desperation for a replacement for HL7v3 among agencies such as ONC. Most of the hype has been created by people with high optimism but a naive understanding of the constraints and difficulties in which e-Health technologies are trying to work.
My recommendation here is simply to carefully consider your requirements and try to understand the paradigms you want to employ in solving them – e.g. whether you want to use an approach such as a semantically-enabled platform architecture, a turn-key commercial solution, or some other approach. Then learn what the scope of various available technologies – what do they address, what do they not address? Any rational person knows that no single technology solves all problems and this is true for both HL7 FHIR and openEHR.
Can openEHR and FHIR work together?
There are trivial and non-trivial answers to this question. If the requirement is making the data in major commercial legacy systems available to applications and to do certain kinds of B2B communications, FHIR is attractive because that’s exactly the problem it was designed to solve, and it may be all that is needed.
However, if the need is to build a long-lived open health platform infrastructure, where the procuring organisation wants control over the data, major component interfaces, and over purchasing choices and timing, a lot more will be needed. In this view of things, EHR isn’t a product you buy, it’s a capability you create, to manage data you will own.
This scenario is what openEHR was designed for – to be part of an open platform infrastructure, as described in the openEHR White Paper. Usually this kind of environment includes major legacy systems and EMRs (this is quite country-dependent), so one of the main jobs an openEHR solution has to perform is implementing data bridges and message import, so as to convert the required data sources to high quality model-based semantic data.
The next challenge is how to implement APIs on the platform in order to enable application building. Historically, the openEHR view of APIs has been closer to those of IHE and OMG services, for example, IHE PIX, PDQ, ATNA and OMG EIS and RLUS. In recent times we have been working on lightweight REST interfaces, both at specification level and with tools such as EhrScape. These provide the native APIs and messaging interfaces of an openEHR environment.
If a procuring authority wants to implement FHIR on top of an openEHR environment in addition to the native interfaces, this will take some effort. There are two approaches. The first is the naive one: essentially the same as implemented an HL7v2 or CDA export channel from the system, which is to say for each FHIR resource+API required, build a custom interface and a back-end that uses AQL to obtain data from the openEHR system. The costs of this are open-ended, since every new API requires more work, but is likely to be quite sustainable for a lot of users.
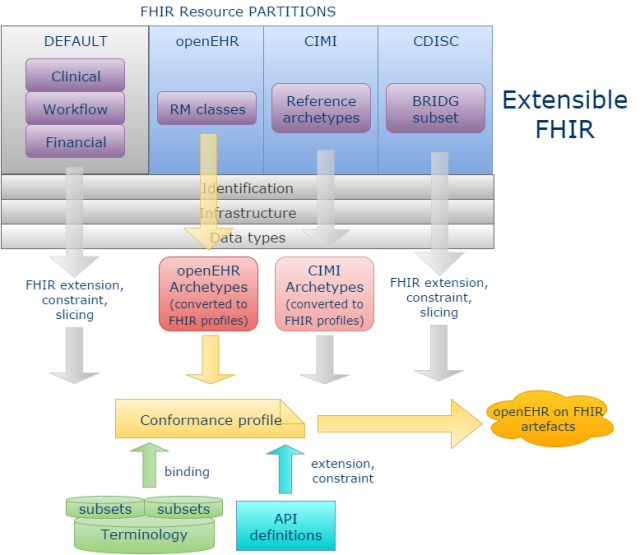
A more powerful and scalable approach would be to enable new ‘resource partitions’ on the generic FHIR communications platform, for example to accommodate the content definitions of openEHR, ISO 13606, CDISC or any other open content model space. This approach is described in detail in the post Making FHIR work for Everybody, from a few months ago (which it has to be said, raised fairly solid resistance within HL7). The essential power of this approach is that is enables programmers to talk to openEHR (or ISO 13606, or CDISC BRIDG …) in the FHIR style, and using the FHIR application development approach, but with the interface content being machine-derived from the relevant source content models rather than the native FHIR resources – the best of both worlds.
My personal belief is that this represents the best hope of bringing these worlds together, but it remains to be seen whether HL7 can be convinced of the value of the idea.
Hopefully this post provides some useful background to those trying to understand openEHR and FHIR, and especially on how they might be used together.
Coda (added 30/Jan)
I realised that a germane thought implied earlier in my discussion should have been included at the end, which is as follows: I believe the main challenge to us working in e-health, and in fact in any domain that has inherent semantic complexity, remains the development of a shared universal library of models of those semantics. Again, the word ‘models’ here means the wide definition – any formalised expression of domain semantics, but expressed distinct from specific time-bound implementation technologies or products.
Some will say that this is impossible, because people won’t agree. But there are levels of agreement. Consider that ‘models’ are really formalisations of theories of some phenomenon (as per this post). Theories start out in the minds of only a few, are discussed, developed, tested, recast, and so on, until they either die or the survive, but in the minds of many. Models are just expressions of theories of understanding, and the level of agreement reflects how widely that understanding is shared.
Where healthcare computing needs to go is a complete separation of models of semantics of healthcare and the technologies used to implement solutions at any given time.



A presentation on HL7 FHIR and openEHR given by FHIR Product Director, Grahame Grieve, at the HL7 FHIR Developer Days Event held in November 2016..
https://vimeopro.com/user12740828/hl7-fhir-developer-days-2016-amsterdam/video/191939835
Interesting presentation – well worth a watch for those interested in this topic. Contains a few inaccuracies re: interoperability (i.e. the idea that openEHR somehow doesn’t do interoperability), but nevertheless a very useful overview of the terrain.
Great overview. I will point colleagues to it as I have got a couple of questions on this topic recently. I do need a couple of more readings of it. However, quick (stupid?) question: Will defined archetypes get identifiers? Can I reference an archetype? Can I look-it up?
@kerfors – this may help – see the Identification and versioning spec on this page – http://www.openehr.org/releases/AM/latest/docs/index
It’s just great to have this omnipresent question answered in such a consistent way. It’s also a typical question you get on every OpenEhr “roadshow” – so this is the must reading for everybody just slightly participating on any OpenEhr pro-motive event.
translated to Japanese. http://openehr.jp/projects/translation-jp/wiki/FHIR%E3%81%A8openEHR%E3%81%AE%E9%81%95%E3%81%84%E3%81%AB%E3%81%A4%E3%81%84%E3%81%A6
This Master thesis compare FHIR and OpenEHR http://ki.se/sites/default/files/eneimi_allwell_brown_a_comparative.pdf
Thanks for the link.
Permanent link to Chinese (zh-CN) translation of this helpful and thoughtful post:
http://mp.weixin.qq.com/s/09unQzlIbebqjtcxnO8n0w
Thanks for your amazing blog 🙂
Best Regards,
Lin Zhang
Pingback: Clinical Modelling – a primer – Lunaria Clinical
Pingback: Global Healthcare Data Standards — A Matter of Time? – Verge HealthTech Fund